Multi-site, Multi-domain Airway Tree Modeling

NEWS
Our Website is permanently transfered to Grand-Challenge.org Page as one of the MICCAI 2022 Challenge Event!
2022/05/30: The Training data have been released! Please refer to Grand-Challenge Page for more details!
2022/05/20: Registration has been open! Please refer to Grand-Challenge Page for more details!
2022/05/01: Official Website Open, Please refer to Grand-Challenge Page for participation.
2022/04/19: Personal Challenge Website Open.
Home
Background and Clinical Significance
Airway segmentation is a crucial step for the analysis of pulmonary diseases including asthma, bronchiectasis, and emphysema. The accurate segmentation based on X-Ray computed tomography (CT) enables the quantitative measurements of airway dimensions and wall thickness, which can reveal the abnormality of patients with chronic obstructive pulmonary disease (COPD). Besides, the extraction of patient-specific airway models from CT images is required for navigation in bronchoscopic-assisted surgery. Due to the fine-grained pulmonary airway structure, manual annotation is however time-consuming, error-prone, and highly relies on the expertise of clinicians.
Objective
Bruijne et al. [1] held the ‘Extraction of airways from CT (EXACT'09)’ challenge in 2009 and achieved a great contribution to the field of airway segmentation. They focused on semi-automated and automated algorithms mainly based on multi-threshold, template matching, and region growing, aiming to relieve the burden of manual delineation and help clinicians explore the influence of pneumonia on airways. These traditional algorithms face difficulty in extracting small peripheral bronchi and suffer the risk of airway leakage. With the advance of deep learning methods, fully convolutional networks (FCNs) achieved state-of-the-art performance in the segmentation task of volumetric medical data. Most of the deep learning methods are data-driven while the EXACT’09 contains only 20 CT scans for training and 20 CT scans for testing, which is not sufficient for the artificial intelligence era. We collected 500 CT scans from multi-sites. The airway tree structures are carefully labeled by three radiologists with more than five years of professional experience. The intra-class imbalance among the trachea, main bronchi, lobar bronchi, and distal segmental bronchi affects the segmentation performance of peripheral bronchi. In conclusion, we encourage the participating teams to design robust algorithms, which can extract the airway tree structure with high topological completeness and accuracy for clinical use. Our challenge is open call (challenge opens for new submissions after conference deadline).
Timeline
20 May 2022 : Registration open.
30 May 2022 : Release of the training and validation data.
15 July 2022 : Open for validation submission.
10 Aug 2022 : Deadline of validation submission and Testing set opening.
25 Aug 2022 : Deadline for testing submission.
18 Sep 2022 : Top 10 teams’ results have been announced.
Organizers
Prof. Guang-Zhong Yang (Institute of Medical Robotics, Shanghai Jiao Tong University)
Dr. Yun Gu (Institute of Medical Robotics, Shanghai Jiao Tong University) Primary Contact
Mr. Minghui Zhang (Institute of Medical Robotics, Shanghai Jiao Tong University) Contact
Mr. Yangqian Wu (Institute of Medical Robotics, Shanghai Jiao Tong University)
Mr. Hanxiao Zhang (Institute of Medical Robotics, Shanghai Jiao Tong University)
Mr. Weihao Yu (Institute of Medical Robotics, Shanghai Jiao Tong University)
Dr. Yulei Qin (Institute of Medical Robotics, Shanghai Jiao Tong University)
Prof. Jiayuan Sun (Department of Respiratory Endoscopy, Shanghai Chest hospital)
Dr. FangFang Xie (Department of Respiratory Endoscopy, Shanghai Chest hospital)
Dr. Xiaoxuan Zheng (Department of Respiratory Endoscopy, Shanghai Chest hospital)
Dataset
500 CT scans (300 for training, 50 for validation, 150 for testing) are collected from multi-sites. The thoracic CT scans are collected from the public LIDC-IDRI dataset [2] and the Shanghai Chest hospital. Each thoracic CT scan is first preprocessed by some strong deep learning models [3][4][5] and ensemble strategy to acquire the preliminary segmentation result and then carefullt delineated and double-checked by three radiologists with more than five years of professional experience to acquire the final refined airway tree structure.
Please refer to Dataset Details to be more familiar with the
dataset structure, information, annotation details, etc,.
Citation: If using this dataset, you must cite the following papers[3-6] . In addition, transfer of the dataset to others is not allowed! The challenge data and results will be free to use after the challenge results publication has been released.
The organizers promise the blackout period will not be no longer than six months after the challenge is over.
Submission Guideline
Please send the signed document to IMR-ATM22@outlook.com for paticipation.
After investigatation, organizing committee will send the access of the data to the teams.
All the registered teams should make a complete submission, containing:
- The docker container. The format of the docker file should follow: Docker Submission Rule.
- A short paper, 2 pages at least, no more than 8 pages. Template same as MICCAI: LNCS.
Evaluation
We aim to segment the accurate airway tree model. For one thing, only the largest component of the binary airway segmentation results are of clinical, to measure the completeness and the connectedness of the participating teams’ results, we use the tree length detected rate (TD) and the branches detected rate (BD). For another thing, we use the metric of the Dice similarity coefficient and Precision to measure the overlap-wise segmentation accuracy. Hence, we finally choose the following metrics:
[1] Tree length detected rate (TD) and the branches detected rate (BD).
[2] Dice similarity coefficient (DSC) and Precision.
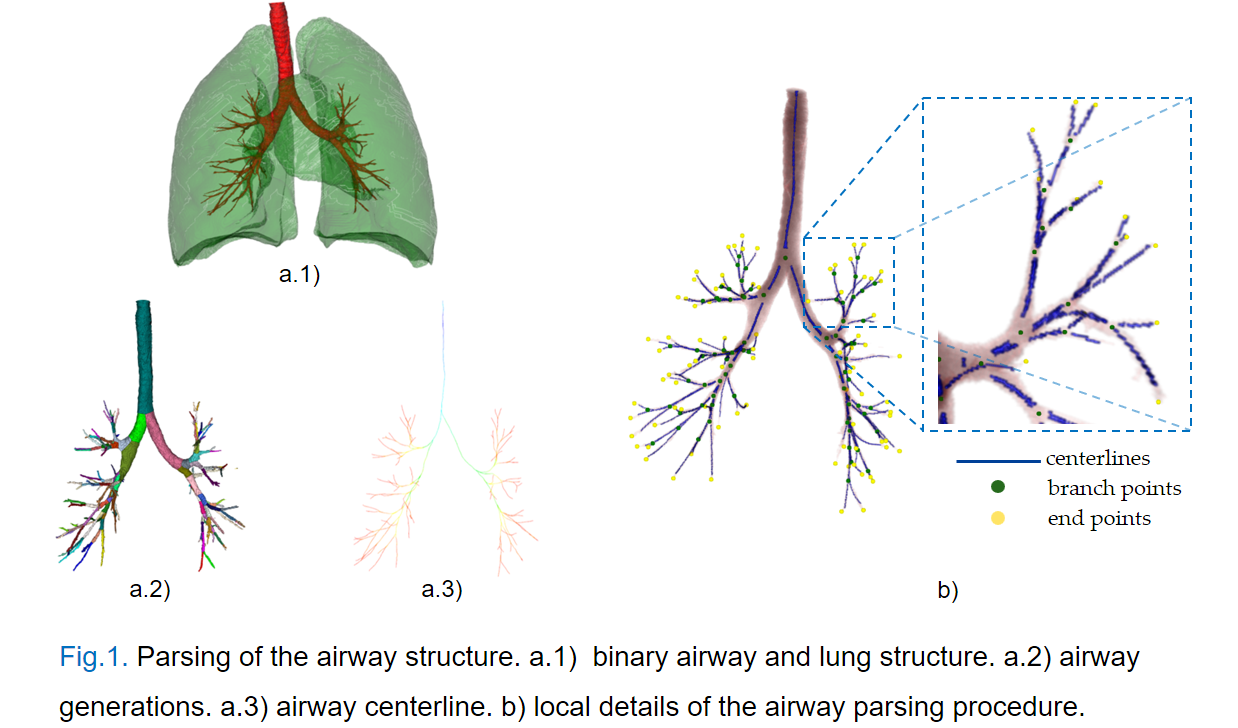
Specifically, The TD is the fraction detected correctly relative to the total tree length in the reference label, and the BD is the percentage of branches that are detected correctly with respect to the total number of branches present in the reference label, as Fig.1 shows the parsing of the airway structure. The above metrics are commonly used in airway segmentations since the EXACT'09 Challenge [1].
Contact Us
If you have any questions about our challenge, please feel free to contact us.
Official Email: IMR-ATM22@outlook.com | Primary Contact: geron762@sjtu.edu.cn | Contact: minghuizhang@sjtu.edu.cn
Reference
Brief survey on airway segmentation: IMR@SJTU_Airway_Segmentation_Survey.pdf .
[1] Lo P, Van Ginneken B, Reinhardt J M, et al. Extraction of airways from CT (EXACT'09)[J]. IEEE Transactions on Medical Imaging, 2012, 31(11): 2093-2107.
[2] Armato III S G, McLennan G, Bidaut L, et al. The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans[J]. Medical physics, 2011, 38(2): 915-931.
[3] Zheng H, Qin Y, Gu Y, et al. Alleviating class-wise gradient imbalance for pulmonary airway segmentation[J]. IEEE Transactions on Medical Imaging, 2021, 40(9): 2452-2462.
[4] Zhang M, Yu X, Zhang H, et al. FDA: Feature Decomposition and Aggregation for Robust Airway Segmentation[M]//Domain Adaptation and Representation Transfer, and Affordable Healthcare and AI for Resource Diverse Global Health. Springer, Cham, 2021: 25-34.
[5] Yu W, Zheng H, Zhang M, et al. BREAK: Bronchi Reconstruction by gEodesic transformation And sKeleton embedding[C]//2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI). IEEE, 2022: 1-5.
[6] Qin Y, Chen M, Zheng H, et al. Airwaynet: a voxel-connectivity aware approach for accurate airway segmentation using convolutional neural networks[C]//International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2019: 212-220.